CRISPR is making headway. The first medical therapy using CRISPR has just been approved. CRISPR-Cas systems show up in complex cells. There’s a lot going on in the genome editing field. Amber Dance wrote an interesting article on these developments in Knowable Magazine.
By accident
Just like many discoveries, CRISPR-Cas was found by accident. In Danisco USA Inc., microbiologists found that bacteria with which they made yoghurt and cheese, were infected with viruses. But when they delved into the matter, they found that some bacteria possessed a defense system to fight off such viral invaders. Consisting of bits of DNA, akin to those in viruses, that bacteria had ‘saved’ in their own genomes. This immune system they dubbed CRISPR-Cas. This shows a viral genome; if the bacterium would encounter the same kind of virus again, it would shred it.
With some modifications, the CRISPR system proved a perfect tool for designing just any genetic sequence. This greatly eased the challenge of genetic engineering. The discovery proved so powerful that it led to a Nobel Prize. And it opened up a new era of gene therapy. Not just in bacteria but also in humans. In December 2023, the US Food and Drug Administration approved the first CRISPR-based gene-changing treatment. A new gene therapy for the excruciatingly painful blood disorder sickle cell anemia.
A precise instrument
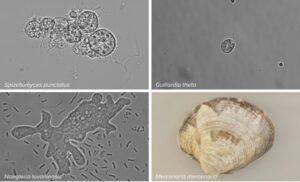
But the development didn’t stop there. Scientists wondered whether similar gene-altering systems also occur in animals, plants and fungi. Life forms with a cell nucleus just like humans. We now know that the answer is yes. In June 2023, a team of MIT and Harvard published a paper in which they showed that CRISPR-like DNA snippers, which they called Fanzors, could be found in different living beings, including fungi, algae, amoebae and a clam called the northern quahog. A discovery that raises many more questions. The most important being: can Fanzors do the same as CRISPR? And might this be the tip of an iceberg of DNA-slicing systems?
The special trick that CRISPR systems offer is to make those cuts only in precise, targeted spots. This requires two elements: a guide to that location, and an enzyme to act as the ‘scissors’. Microbes have evolved a handful of scissor enzymes that make this cut, with names like Cas9 and Cas12. When microbes are infected by viruses, the microbes collect small pieces of the viral genetic sequences and stow them together in a segment of their genome called CRISPR repeats. The next time that virus comes along, the microbe can use those sequences to create the guide; the enzyme scissors snip up the genetic material of the virus and defend itself. About half of all known bacteria, as well as most of other microbes known as archaea, have CRISPR-Cas systems. But CRISPR-Cas can also be used to repair broken DNA strands. Either imperfectly (possibly leading to new disorders) or perfectly.
A universal system
Every living organism uses the same basic DNA code and proteins, so the CRISPR-Cas system can theoretically work in any organism scientists drop it into — though in reality, some tweaking is generally required. Using a few extra molecular tricks, researchers can also use CRISPR to add a novel genetic sequence or to repair a broken gene. In this way, scientists can use CRISPR-Cas to treat diabetes; a form of amyloidosis; infections, including HIV; and a variety of cancers. And that’s not all. Researchers are applying CRISPR to turn pigs into possible organ donors for people, create better-quality, disease-resistant fruits, and eradicate mosquitoes that transmit malaria.
But there are downsides. Breaking a strand of DNA in people is a risky business: If Cas cuts in the wrong place or the repair process goes wrong, the therapy could change the genome in a way that triggers cancer. Thus, many modern CRISPR-based approaches use modified versions of the Cas protein that don’t fully cut the DNA but edit it more subtly and safely.
Fanzor
Now about Fanzor. This has been ‘hiding in plain sight,’ according to David J. Segal, a geneticist at UC Davis who wrote about the new age of genome engineering for the Annual Review of Genomics and Human Genetics in 2013. The Fanzor genes were first described that same year, but no one knew they encoded a Cas-like enzyme.
The discovery of Fanzor resulted from a study on ‘jumping genes’ – DNA sequences that can jump out of one spot in the genome and reinsert themselves somewhere else. Weidong Bao, a researcher at the Genetic Information Research Institute in Cupertino, California, knew that bacterial jumping genes often contain a gene of unknown function called TnpB. He went looking for similar genes and found them in the genomes of more than two dozen eukaryotes, including a fly, yeasts and molds, amoebas and several algae. The researchers named the eukaryotic version of this mysterious gene Fanzor instead of TnpB. In later research into this phenomenon, it was discovered that Fanzor is a cousin of the bacterial Cas genes; both gene families were descended from TnpB.
Not very specific
It has been proven by Zhang and colleagues that Fanzor genes carry instructions for cutting DNA. But such an indication is still a long way of using Fanzor for cutting DNA at specific sites. Up till now, the best Zhang’s team has been able to do is to cut the target gene site 18.4% of the time. Still, they haven’t even tried to repair a faulty gene with it. That’s not nearly as good as CRISPR-Cas, which has been developed over the years.
Fanzor has some advantages and disadvantages over CRISPR-Cas. It is smaller, and can therefore better be delivered to the DNA in question. On the other hand, scientists have already designed miniature versions of the Cas12 and Cas 13 enzymes that can fit into viruses. So that’s a bit of a draw between the two. Then, there is the problem that Fanzor is shorter than CRISPR-Cas: 15 DNA bases versus 18 to 20. This means that Fanzor might indicate a place to cut other than the target site. If it would also cut other sites, this could be disastrous to Fanzor gene therapy.
So why change?
So why change? CRISPR works ‘just fine in most organisms,’ says Sophien Kamoun, a plant pathologist at the Sainsbury Laboratory in Norwich in the United Kingdom who has reviewed the use of CRISPR to engineer crop plants. But Fanzor might still be useful for scientists working with certain bacteria where Cas enzymes are toxic, suggests Kamoun. Still, Fanzor is a very cool biological discovery.
While we know what CRISPR-Cas systems do for microbes, it’s not clear exactly what Fanzor proteins do in nature. Researchers believe they’re somehow partnered with jumping genes — maybe helping them, or perhaps hitching a ride.
One organism that doesn’t naturally have Fanzors, Zhang says, is humans. But maybe, people possess some other, similar DNA-cutting system in the Fanzor-Cas family? Zhang’s group is actively hunting for new ones. He says that they try to discover as many DNA-cutting systems as possible.
Interesting? Then also read:
CRISPR-Cas: a prize winning technology?
Duchenne muscular dystrophy CRISPR cure: just for the rich and well-connected?
CRISPR to the rescue as bacteria develop antibiotics resistance
